Medical Named Entity Recognition using Fine-Tuned BERT and ALBERT
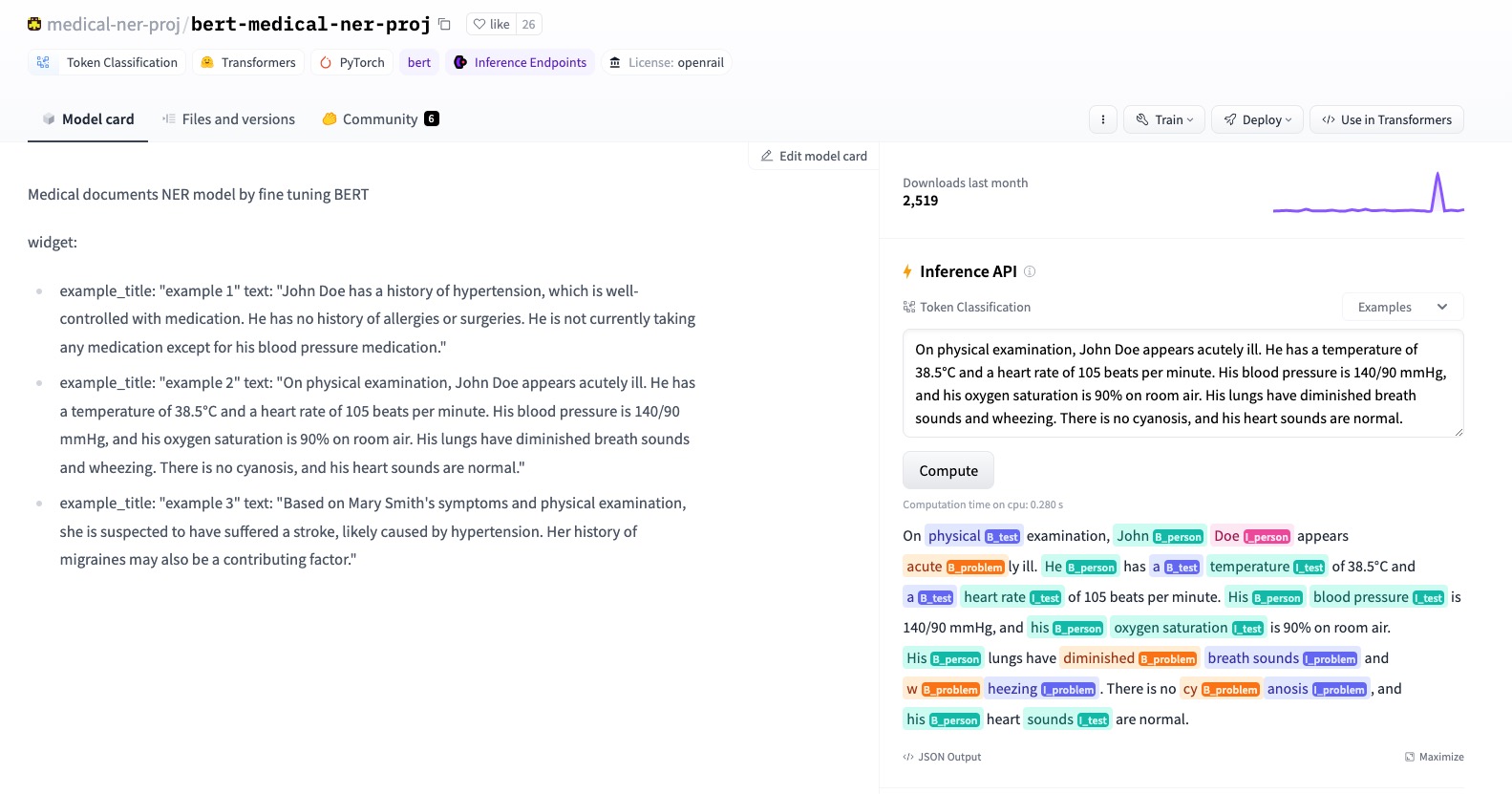
Project Overview: This repository is dedicated to advancing Medical Named Entity Recognition (NER) using cutting-edge pre-trained models such as BERT and ALBERT. By fine-tuning these models on the i2b2/n2c2 dataset, which contains annotated clinical notes, the project evaluates their performance in identifying medical entities, contributing significantly to the field of medical informatics.
Fine-tuned LLM Models : BERT | ALBERT
Objectives
- Fine-tune pre-trained BERT and ALBERT models on the i2b2/n2c2 dataset for medical NER tasks.
- Evaluate and compare the performance of these models in recognizing medical entities from clinical notes.
- Make the fine-tuned models accessible for further research and application through Huggingface.
Features
- Pre-trained Models Fine-Tuning:
- Utilizes BERT and ALBERT, two of the most advanced pre-trained models, and fine-tunes them for Medical NER tasks.
- Comprehensive Dataset:
- Employs the i2b2/n2c2 dataset, which is rich in annotated clinical notes, for training and evaluating the models.
- Detailed Evaluation:
- Assesses the models’ performance using metrics such as F1-score and accuracy, providing a thorough comparison between the two.
- Huggingface Deployment:
- The fine-tuned models are uploaded to Huggingface, making them available for public use and further research.
Technology Stack
- Programming Languages: Python 3.x for model training and evaluation.
- Libraries and Frameworks: Jupyter Notebook for interactive development, Huggingface Transformers for model architecture, and PyTorch for deep learning operations.
- Data Processing: Custom scripts for parsing the i2b2/n2c2 dataset and preparing it for NER tasks.
Outcome
The project successfully demonstrates the effectiveness of fine-tuning pre-trained BERT and ALBERT models for the task of Medical Named Entity Recognition. Key outcomes include:
- Enhanced Model Performance: Both BERT and ALBERT models showed excellent capability in recognizing medical entities, achieving high F1-scores and accuracy on the i2b2/n2c2 dataset.
- Model Accessibility: Fine-tuned models have been made available on Huggingface, providing a valuable resource for the research community and practitioners in the medical informatics field.
- Practical Implications: The project’s findings highlight the potential for applying advanced NLP techniques to improve information extraction from clinical notes, aiding in patient care and medical research.
Learning
This project offered an invaluable learning experience in several key areas:
- Advanced NLP Techniques: Gained insights into the process of fine-tuning pre-trained language models for specialized tasks such as NER.
- Medical Informatics: Enhanced understanding of the challenges and opportunities in applying NLP to the medical domain, especially in processing and extracting meaningful information from clinical texts.
- Technical Skills: Developed proficiency in using Python, Jupyter Notebook, Huggingface Transformers, and PyTorch, along with skills in data parsing and preprocessing for NLP tasks.
Usage
- Data Parsing: Run
parsing_n2c2_to_conll.ipynbto parse the dataset. - BERT Fine-tuning: Execute
bert_finetuning.ipynbfor BERT model fine-tuning. - ALBERT Fine-tuning: Execute
albert_finetuning.ipynbfor ALBERT model fine-tuning.
Models will be saved in their respective folders upon completion.
Results
- BERT: Achieved an F1-score of 0.8726 and an accuracy of 0.9557.
- ALBERT: Attained an F1-score of 0.8667 and an accuracy of 0.9518.
For a detailed breakdown of results, refer to the included Report.
References
This project draws upon a wide array of research and resources in the domain of NER and medical informatics, including BioELECTRA and various Huggingface guides.
Note: This project documentation is intended to guide developers, researchers, and enthusiasts interested in applying deep learning techniques to the medical domain, specifically for Named Entity Recognition tasks.